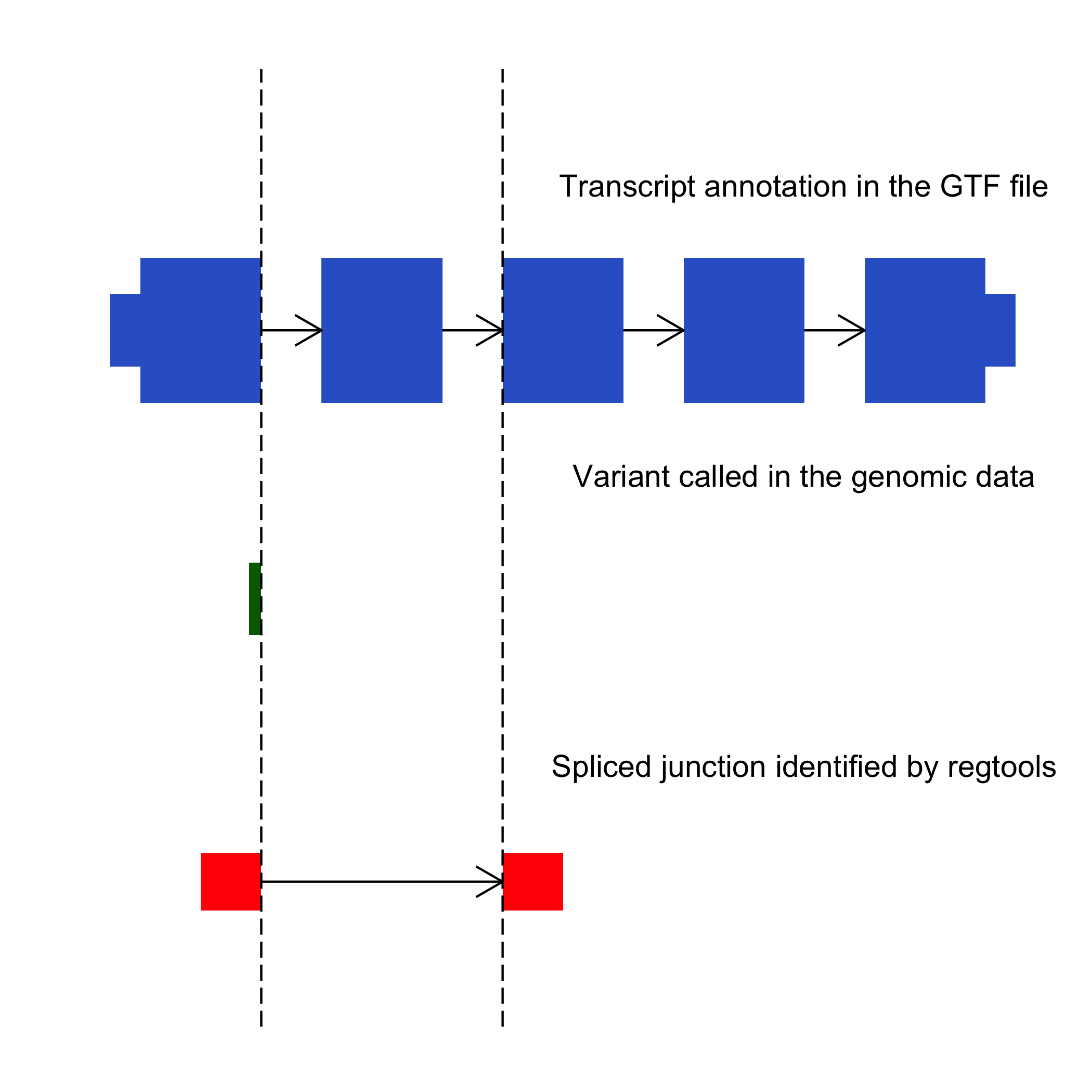
The cis-splice-effects identify command is used to identify splicing misregulation events. This command takes in a list of variants in the VCF format and RNAseq alignments produced with a splice-aware aligner in the BAM format. The tool then proceeds to identify non-canonical splicing junctions near the variant sites.
Usage
regtools cis-splice-effects identify [options] variants.vcf alignments.bam ref.fa annotations.gtf
Input
| Input | Description |
|---|---|
| variants.vcf | Variant call in VCF format from which to look for cis-splice-effects. |
| alignments.bam | Aligned RNAseq BAM/CRAM produced with a splice aware aligner, that has been indexed for example with samtools index. We have tested this command with alignments from HISAT2, TopHat2, STAR, kallisto, and minimap2. |
| ref.fa | The reference FASTA file. The donor and acceptor sequences used in the "splice-site" column of the annotated junctions are extracted from the FASTA file. |
| annotations.gtf | The GTF file specifies the transcriptome that is used to annotate the junctions and variants. For examples, the Ensembl GTFs for release 106 are here. |
Note - Please make sure that the version of the annotation GTF that you use corresponds with the version of the assembly build (ref.fa) and that the co-ordinates in the VCF file are also from the same build.
Options
| Option | Description |
|---|---|
| -o STR | Output file containing the aberrant splice junctions with annotations. [STDOUT] |
| -v STR | Output file containing variants annotated as splice relevant (VCF format). |
| -j STR | Output file containing the aberrant junctions in BED12 format. |
| -s INT | Strand specificity of RNA library preparation, where the options XS, use XS tags provided by aligner; RF, first-strand; FR, second-strand. This option is required. If your alignments contain XS tags, these will be used in the "unstranded" mode. If you are unsure, we have created this table to help. |
| -w INT | Window size in b.p to identify splicing events in. The tool identifies events in variant.start +/- w basepairs. Default behaviour is to look at the window between previous and next exons. |
| -e INT | Maximum distance from the start/end of an exon to annotate a variant as relevant to splicing, the variant is in exonic space, i.e a coding variant. [3] |
| -i INT | Maximum distance from the start/end of an exon to annotate a variant as relevant to splicing, the variant is in intronic space. [2] |
| -I | Annotate variants in intronic space within a transcript(not to be used with -i). |
| -E | Annotate variants in exonic space within a transcript(not to be used with -e). |
| -S | Don't skip single exon transcripts. |
| -b | The file containing the barcodes of interest for single cell data. |
| -C | Tells cis-splice-effects identify that you want intron-motif method to take priority when assigning strand. i.e. decide strandedness based on the fasta rather than what is encoded in the alignment file. |
Note Both junctions extract and cis-splice-effects identify have an intron-motif method that can be used to determine strandedness of junctions extracted from alignment files. Using this method supercedes any strandedness information that might be encoded in the alignment file. To use this method with cis-splice-effects identify, you can the -C option to tell RegTools that you want the intron-motif method to take priority when assigning strandedness.
Output
For an explanation of the annotated junctions that are identified by this command please refer to the output of the junctions annotate command here
For an explanation of the annotated variants that are identified by this command when using the -v option, please refer to the output of the variants annotate command here
Examples